Robust, user-friendly, free, open-source Software: openGUTS
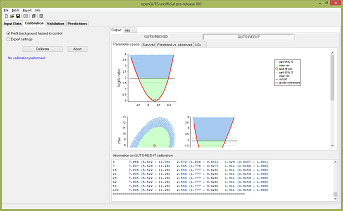
OpenGUTS is a user-friendly software to perform analyses with GUTS: the General Unified Threshold model for
Survival. GUTS is the leading toxicokinetic-toxicodynamic framework for the endpoint survival (and other non-reversible all-or-nothing effects).
This site
hosts the software itself, as a standalone Windows executable, with manuals and background documentation. Since openGUTS is (as the name implies) open source, also the source files can be
obtained here. Additionally, you can download the Matlab version, which served as the
blueprint for the standalone version. The Matlab version has basically the same functionality, and can be easily adapted or extended to perform customised analyses. Finally, we provide (links to)
further information on openGUTS and GUTS. The main features of the software are:
- Open and free: the software is open source and freely downloadable. The fully-functional Matlab version that served as the prototype is also available.
- User-friendly: fit models and derive confidence intervals without requiring user interaction (e.g., no need for starting values).
- Robust: always find the global optimum and relevant intervals, even for awkward data sets.
- Flexible: allow time-varying exposure, missing data, simultaneous fitting on multiple data sets, etc.
- Efficient: rapid screening of exposure profiles (e.g., FOCUS output) by batch processing.
- Supportive: the software follows the workflow as laid down in the 2018 EFSA opinion on TKTD models.
Funginol dataset for openguts exercises
These files can be loaded into openGUTS to do model calibration, validation and environmental risk assessment of time-variable exposure with different aquatic exposure profiles.
openguts website
The openGUTS website is now the best starting point to learn about GUTS software.
EFSA scientific opinion on TKTD models
The EFSA scientific opinion on TKTD modelling has been published! Read here...
Book: Modelling survival under chemical stress

Survival is a relevant endpoint for many questions related to the effects of chemicals in the environment. Making sense of mortality, as a process over time, requires mechanism-based models, known as toxicokinetic-toxicodynamic (TKTD) models. All published TKTD models for survival can now be viewed as members of an over-arching framework: GUTS.
Click on the book cover or here to view the e-book.
Preface
1 Introduction
2 Description of GUTS
3 Mathematical treatment
4 Case study: dieldrin in guppies
5 Case study: propiconazole in amphipods
6 Use cases
7 Ring test
8 Model evaluation
9 Outlook
Bibliography
Glossary
Appendices
Modelling survival: exposure pattern, species sensitivity and uncertainty

The General Unified Threshold model for Survival (GUTS) integrates previously published toxicokinetic-toxicodynamic models and estimates survival with explicitly defined assumptions. Importantly, GUTS accounts for time-variable exposure to the stressor. We performed three studies to test the ability of GUTS to predict survival of aquatic organisms across different pesticide exposure patterns, time scales and species. Firstly, using synthetic data, we identified experimental data requirements which allow for the estimation of all parameters of the GUTS proper model. Secondly, we assessed how well GUTS, calibrated with short-term survival data of Gammarus pulex exposed to four pesticides, can forecast effects of longer-term pulsed exposures. Thirdly, we tested the ability of GUTS to estimate 14-day median effect concentrations of malathion for a range of species and use these estimates to build species sensitivity distributions for different exposure patterns. We find that GUTS adequately predicts survival across exposure patterns that vary over time. When toxicity is assessed for time-variable concentrations species may differ in their responses depending on the exposure profile. This can result in different species sensitivity rankings and safe levels. The interplay of exposure pattern and species sensitivity deserves systematic investigation in order to better understand how organisms respond to stress, including humans. (open access, Nature Scientific Reports)
old Software implementations of GUTS (outdated!)
Warning: These implementations are outdated. It is much better to use openGUTS!
The zipped folder contains a R script, an executable programmed in Delphi, MATLAB files, ModelMaker files and Mathematica implementations of GUTS. This software is made available under the GNU General Public License 3.0. The software contains errors and bugs. We do not accept liability or responsibility for any damage or costs incurred as a result of these errors.
Death Dilemma and organism recovery in ecotoxicology

Why do some individuals survive after exposure to chemicals while others die? Either, the tolerance threshold is distributed among the individuals in a population, and its exceedance leads to
certain death, or all individuals share the same threshold above which death occurs stochastically. The previously published General Unified Threshold model of Survival (GUTS) established a
mathematical relationship between the two assumptions. According to this model stochastic death would result in systematically faster compensation and damage repair mechanisms than individual
tolerance. Thus, we face a circular conclusion dilemma because inference about the death mechanism is inherently linked to the speed of damage recovery. We provide empirical evidence that the
stochastic death model consistently infers much faster toxicodynamic recovery than the individual tolerance model. Survival data can be explained by either, slower damage recovery and a wider
individual tolerance distribution, or faster damage recovery paired with a narrow tolerance distribution. The toxicodynamic model parameters exhibited meaningful patterns in chemical space, which
is why we suggest toxicodynamic model parameters as novel phenotypic anchors for in vitro to in vivo toxicity extrapolation. GUTS appears to be a promising refinement of traditional survival
curve analysis and dose response models. (Published in Environmental Science & Technology, open access)
General Unified Threshold Model of Survival - a Toxicokinetic-Toxicodynamic Framework for Ecotoxicology
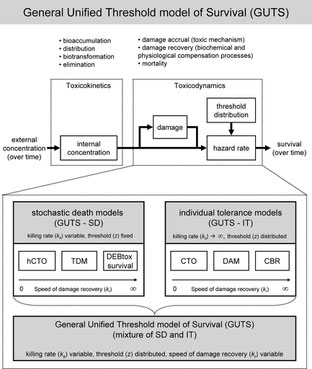
Toxicokinetic-toxicodynamic models (TKTD models) simulate the time-course of processes leading to toxic effects on organisms. Even for an apparently simple endpoint as survival, a large number of very different TKTD approaches exist. These differ in their underlying hypotheses and assumptions, although often the assumptions are not explicitly stated. Thus, our first objective was to illuminate the underlying assumptions (individual tolerance or stochastic death, speed of toxicodynamic damage recovery, threshold distribution) of various existing modeling approaches for survival and show how they relate to each other (e.g., critical body residue, critical target occupation, damage assessment, DEBtox survival, threshold damage). Our second objective was to develop a general unified threshold model for survival (GUTS), from which a large range of existing models can be derived as special cases. Specific assumptions to arrive at these special cases are made and explained. Finally, we illustrate how special cases of GUTS can be fitted to survival data. We envision that GUTS will help increase the application of TKTD models in ecotoxicological research as well as environmental risk assessment of chemicals. It unifies a wide range of previously unrelated approaches, clarifies their underlying assumptions, and facilitates further improvement in the modeling of survival under chemical stress. (link at journal) or (download here)
More information on GUTS
More information on GUTS, including Matlab code, can be found on Tjalling Jager's website. Furthermore there is also a GUTS r-package available.
Use of the special cases GUTS-SD and GUTS-IT
Using the General Unified Threshold model of Survival this study systematically investigates how well different ways of using the model perform when it comes to predicting survival: "Toxicokinetic-toxicodynamic modelling of survival of Gammarus pulex in multiple pulse exposures to propiconazole: model assumptions, calibration data requirements and predictive power." Ecotoxicology (link to paper, free access)
Toxicokinetic-toxicodynamic (TKTD) models quantify the time-course of internal concentration, which is defined by uptake, elimination and biotransformation (TK), and the processes which lead to the toxic effects (TD). TKTD models show potential in predicting pesticide effects in fluctuating concentrations, but the data requirements and validity of underlying model assumptions are not known. We calibrated TKTD models to predict survival of Gammarus pulex in propiconazole exposure and investigated the data requirements. In order to assess the need of TK in survival models, we included or excluded simulated internal concentrations based on pre-calibrated TK. Adding TK did not improve goodness of fits. Moreover, different types of calibration data could be used to model survival, which might affect model parameterization. We used two types of data for calibration: acute toxicity (standard LC50, 4 d) or pulsed toxicity data (total length 10 d). The calibration data set influenced how well the survival in the other exposure scenario was predicted (acute to pulsed scenario or vice versa). We also tested two contrasting assumptions in ecotoxicology: stochastic death and individual tolerance distribution. Neither assumption fitted to data better than the other. We observed in 10-d toxicity experiments that pulsed treatments killed more organisms than treatments with constant concentration. All treatments received the same dose, i.e. the time-weighted average concentration was equal. We studied mode of toxic action of propiconazole and it likely acts as a baseline toxicant in G. pulex during 10-days of exposure for the endpoint survival.
Computationally Efficient Implementation of a Novel Algorithm for the General Unified Threshold Model of Survival (GUTS)
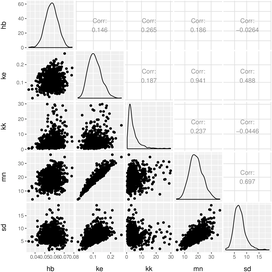
The General Unified Threshold model of Survival (GUTS) provides a consistent mathematical framework for survival analysis. However, the calibration of GUTS models is computationally challenging. We present a novel algorithm and its fast implementation in our R package, GUTS, that help to overcome these challenges. We show a step-by-step application example consisting of model calibration and uncertainty estimation as well as making probabilistic predictions and validating the model with new data. Using self-defined wrapper functions, we show how to produce informative text printouts and plots without effort, for the inexperienced as well as the advanced user. The complete ready-to-run script is available as supplemental material. We expect that our software facilitates novel re-analysis of existing survival data as well as asking new research questions in a wide range of sciences. In particular the ability to quickly quantify stressor thresholds in conjunction with dynamic compensating processes, and their uncertainty, is an improvement that complements current survival analysis methods. PLoS Comput Biol 12(6): e1004978.
